-Search query
-Search result
Showing all 50 items for (author: echeverria & i)
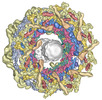
EMDB-41123: 
Composite multiscale structure of the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41114: 
Nuclear double outer ring of the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41116: 
Lumenal ring of the isolated yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41117: 
Pom34-Pom152 membrane attachment site yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41119: 
Cytoplasmic outer ring of the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41120: 
Central transporter/plug yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41121: 
Double outer ring connectors of the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41122: 
Connectors in cytoplasmic outer ring of yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41285: 
Double nuclear outer ring of Nup84-complexes from the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-41300: 
Inner spoke ring of the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

PDB-8t9l: 
Pom34-Pom152 membrane attachment site yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

PDB-8tie: 
Double nuclear outer ring of Nup84-complexes from the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

PDB-8tj5: 
Inner spoke ring of the yeast NPC
Method: single particle / : Akey CW, Echeverria I, Ouch C, Fernandez-Martinez J, Rout MP

EMDB-26574: 
KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

EMDB-27002: 
ACP1-KS-AT domains of mycobacterial Pks13
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

EMDB-27003: 
KS-AT domains of mycobacterial Pks13 with inward AT conformation
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

EMDB-27004: 
KS-AT domains of mycobacterial Pks13 with outward AT conformation
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

EMDB-27005: 
ACP1-KS-AT domains of mycobacterial Pks13
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

PDB-7uk4: 
KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

PDB-8cuy: 
ACP1-KS-AT domains of mycobacterial Pks13
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

PDB-8cuz: 
KS-AT domains of mycobacterial Pks13 with inward AT conformation
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

PDB-8cv0: 
KS-AT domains of mycobacterial Pks13 with outward AT conformation
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

PDB-8cv1: 
ACP1-KS-AT domains of mycobacterial Pks13
Method: single particle / : Kim SK, Dickinson MS, Finer-Moore JS, Rosenberg OS, Stroud RM

EMDB-27032: 
Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

EMDB-27033: 
Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

EMDB-27034: 
Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

EMDB-28667: 
Cryo-EM map of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1-prime
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

PDB-8cx0: 
Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

PDB-8cx1: 
Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

PDB-8cx2: 
Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2
Method: single particle / : Li Y, Langley C, Azumaya CM, Echeverria I, Chesarino NM, Emerman M, Cheng Y, Gross JD

EMDB-24224: 
Combined 3D structure of the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echeverria I, Fernandez-Martinez J, Nudelman I

EMDB-24225: 
Low resolution, multi-part 3D structure of the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echeverria I, Fernandez-Martinez J, Nudelman I

EMDB-24231: 
Double nuclear outer ring from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echevarria I, Fernandez-Martinez J, Nudelman I

EMDB-24232: 
Inner ring spoke from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echevarria I, Fernandez-Martinez J, Nudelman I

EMDB-24258: 
Structure of the in situ yeast NPC
Method: subtomogram averaging / : Villa E, Singh D, Ludtke SJ, Akey CW, Rout MP, Echeverria I, Suslov S
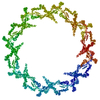
PDB-7n84: 
Double nuclear outer ring from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echevarria I, Fernandez-Martinez J, Nudelman I
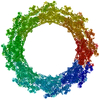
PDB-7n85: 
Inner ring spoke from the isolated yeast NPC
Method: single particle / : Akey CW, Rout MP, Ouch C, Echevarria I, Fernandez-Martinez J, Nudelman I

PDB-7n9f: 
Structure of the in situ yeast NPC
Method: subtomogram averaging / : Villa E, Singh D, Ludtke SJ, Akey CW, Rout MP, Echeverria I, Suslov S

EMDB-11609: 
SARS-CoV-2-Nsp1 bound to a translationally inactive 80S ribosome
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muehlemann O, Ban N

EMDB-11321: 
SARS-CoV-2-Nsp1-40S complex, focused on body
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muehlemann O, Ban N

PDB-6zok: 
SARS-CoV-2-Nsp1-40S complex, focused on body
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muehlemann O, Ban N

EMDB-11320: 
SARS-CoV-2-Nsp1-40S complex, composite map
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut ML, Thiel V, Muehlemann O, Ban N

EMDB-11322: 
SARS-CoV-2-Nsp1-40S complex, focused on head
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut ML, Thiel V, Muehlemann O, Ban N

EMDB-11323: 
SARS-CoV-2-Nsp1 bound to 43S Pre-initiation complex
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muehlemann O, Ban N

PDB-6zoj: 
SARS-CoV-2-Nsp1-40S complex, composite map
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut ML, Thiel V, Muehlemann O, Ban N

PDB-6zol: 
SARS-CoV-2-Nsp1-40S complex, focused on head
Method: single particle / : Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut ML, Thiel V, Muehlemann O, Ban N

EMDB-0071: 
GAPDH-CP12-PRK complex
Method: single particle / : McFarlane CR, Shah N, Bubeck D, Murray JW

PDB-6gve: 
GAPDH-CP12-PRK complex
Method: single particle / : McFarlane CR, Shah N, Bubeck D, Murray JW

EMDB-7321: 
Integrative Structure and Functional Anatomy of a Nuclear Pore Complex
Method: subtomogram averaging / : Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Ludtke SJ, Akey CW, Chait BT, Sali A, Rout MP

EMDB-8696: 
Regulation of Rvb1/Rvb2 by a domain within the INO80 chromatin remodeling complex implicates the yeast Rvbs as protein assembly chaperones
Method: single particle / : Zhou CY, Stoddard CI, Johnston JB, Trnka MJ, Echeverria I, Palovcak E, Sali A, Burlingame AL, Cheng Y, Narlikar GJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
